Let’s get personal – better predicting individual response with more biomarkers
The project that’s changing how we look at biomarkers in cancer research and precision oncology

Biomarkers go hand in hand with cancer biology. They’ve even become somewhat of a buzzword over the years – identifying the best biomarker, using the right biomarker – and it’s warranted. Biomarkers are an integral and essential part of basic, translational and clinical research, helping improve the ability to diagnose, monitor, and predict disease risk and progression.
Once identified and validated, biomarkers can be key in informing disease status (such as with HER2) or predicting therapeutic efficacy (like BRAF mutations can) … but finding the right biomarker that answers the question being asked can be difficult. The wrong biomarker can cause variability in results. It may be relevant in some cases but not others, confounding its usefulness and constraining it to only certain outcomes. And with therapeutic approaches becoming more complex as we learn more about a disease, finding effective biomarkers can be a significant challenge.
In cancer research, many conventional therapeutic strategies rely on single biomarker analysis, severely limiting input and commonly resulting in a high poor response rate, varying clinical trial outcomes and ineffective target-to-drug matches. Single biomarker strategies tend to unintentionally simplify tumor biology, a complex and evolving ecosystem in which many variables can have an impact.
Bigger biomarker profiles for a more systemic view
What if multiple biomarkers are used? Can a biomarker profile be more accurate in its assessments and predictions? Well, of course – integrating different layers of molecular and cellular information, such as phosphorylation, cell–cell interactions, tumor microenvironment structure, and inflammatory and immune markers, can provide a much clearer and more comprehensive evaluation of how well a drug might work for a specific individual, and can ultimately help advance personalized predictive decision-making.
In fact, incorporating data from a multiple-marker profile using multidimensional data generated from a multimodal technology approach has the potential to uncover novel therapeutic targets and combinations that might have been overlooked if only relying on a single biomarker. Historically, multidimensional data has been time-consuming and expensive to generate – with the need to utilize several different systems, not to mention a lot of precious sample.
Recent advances in functional and omics technologies – especially at the single-cell level – are reshaping clinical oncology research, making multimodal approaches more accessible. These technologies allow for deep molecular profiling of tumors, revealing individual therapeutic vulnerabilities based on a larger biomarker set. As we overcome historical barriers and with continued work and progress, application of multi-omic and single-cell data can become more routine in improving disease management.
The feasibility study for multi-omic tumor profiling
A large collaboration based in Zurich is working on this by integrating cutting-edge technologies for the detailed and comprehensive investigation of tumor biopsies in a prospective multicohort precision oncology project. The Tumor Profiler (TuPro) project was developed to assess whether functional, single-cell and bulk omics readouts can be effective in supporting decisions for cancer therapy strategies and to provide information on additional, potentially relevant biomarkers beyond current guidelines and standard of care.
The latest data to come from this ongoing project reports the application of nine technologies to help determine the outcomes for 116 melanoma patients enrolled in the TuPro study. This magnitude of multidimensional data for each patient could enable the in-depth characterization of molecular drivers and therapeutic response specific to an individual tumor, opening new opportunities for personalized treatments and more effective patient stratification.
This groundbreaking study published in Nature Medicine by the TuPro consortium demonstrates the ability to operationalize multi-omics for the development of biomarker profiles that could help guide selection of treatments in melanoma – a cancer known for its heterogeneity and therapeutic resistance.
In advanced melanoma, the introduction of immune checkpoint inhibitors targeting PD-1, CTLA-4 and LAG-3, as well as therapies focusing on dysregulation in the MAPK pathway, has dramatically improved survival rates. Yet, reliance on single biomarkers, which for melanoma is solely BRAF class I mutations, has shown limitations when determining eligibility for treatment options. This underscores the need for broader mechanism-based biomarker panels to enable better therapy prediction based on the tumor biology of individual patients.
The study evaluated the feasibility and utility of multi-omics in generating data that can be used to identify better predictive biomarkers by leveraging a comprehensive suite of omics technologies – including genomics, transcriptomics, proteomics and single-cell analyses – to profile tumors from patients with advanced melanoma. High-quality molecular datasets were generated (up to 500 GB of data per sample, translating to 40,000 potential markers) from 126 melanoma tumor biopsies using two standard (DigiPath and NGS) and seven experimental technologies (pharmacoscopy, 4iDRP, IMC™, CyTOF™, proteotyping, scRNA-seq and scDNA-seq). Unlike traditional approaches that rely on a limited set of biomarkers, this multimodal strategy enabled a more systemic understanding of tumor biology, the role of immune response and potential mechanisms of therapeutic resistance by developing a 54-biomarker profile with which to inform and predict treatment outcomes.
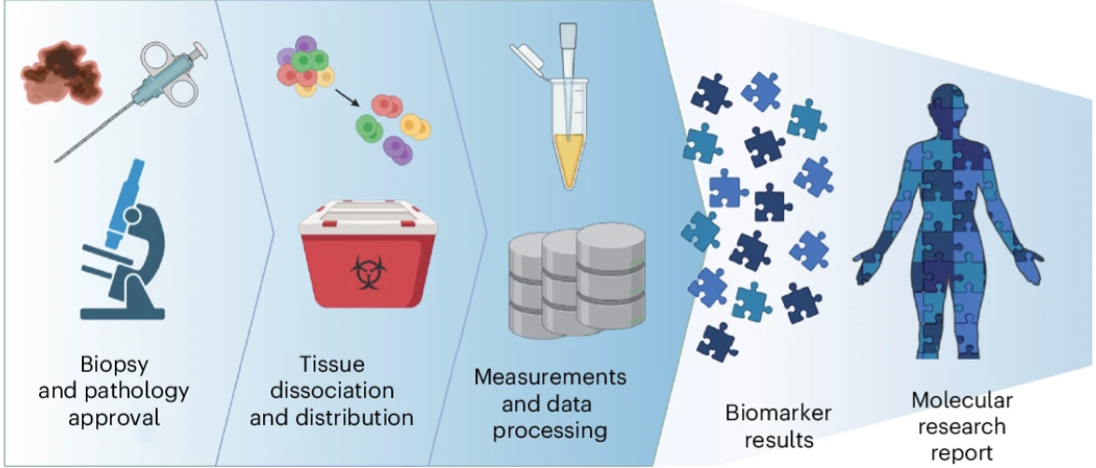
The TuPro biomarker analysis workflow (Miglino, N. et al.)
By combining these technologies into a single analysis pipeline, the team achieved a fast turnaround time from sample acquisition to analysis and reporting of four weeks. This accelerated timeframe demonstrates the viability and practicality of incorporating multi-omic technologies into formulating how future treatment strategies could be applied. These findings highlight the value of integrating high-dimensional data into therapeutic strategy guidelines, particularly in cancers in which standard treatments often fall short.
For clinical and translational research, these results open new avenues of understanding. Multi-omics can accelerate biomarker discovery, refine patient stratification and inform adaptive trial designs. And with the technologies chosen and pipeline developed, key challenges that limit standard clinical trials were also improved, for example, data harmonization, turnaround time and the need for a robust bioinformatics infrastructure.
Moving cancer research forward
The TuPro initiative exemplifies a move toward systems-level thinking in oncology. As multi-omic platforms become more accessible and computational tools more sophisticated, there is a tangible opportunity to translate these insights into enhanced therapeutic development strategies and accelerate precision medicine initiatives that could significantly improve individual patient outcome.
About two of the technologies used in the study
CyTOF and Hyperion™ systems were leveraged for targeted proteomics in this study. The selection of these technologies was based on their capability to provide a comprehensive molecular portrait of each tumor and to analyze limited abundance of sample.
CyTOF | Single-cell proteomics
The CyTOF platform offers high-resolution analysis of over 50 markers in a single assay, generating a massive amount of single-cell data per sample in a short amount of time. Mass cytometry–based CyTOF systems are the most widely published solution for high-parameter immune research, appearing in more than 2,600 peer-reviewed publications and more than 250 clinical trials, with 100% of the top 10 cancer immunotherapies leveraging unique biomarkers found by mass cytometry.
Hyperion | Spatial proteomics
The Hyperion XTi Imaging System can detect 40-plus protein markers across a broad dynamic range with the highest throughput. Powered by IMC technology, the platform uniquely empowers researchers to mix and match different antibodies without extensive assay validation and enables the quantitation of weak signals in the presence of bright signals in the same sample.
