Multiple myeloma and IMC technology
How spatial biology supports multiple myeloma research
Spatial biology … the term is seemingly everywhere and integrated into all the latest studies – as it should be. Visualizing data in a dimensional context can be an important asset to understanding how and why cells behave as they do. Detailed insights into cellular initiation and progression can in turn help us determine how we can prevent, treat and possibly even cure diseases.
So, what is spatial biology? Spatial biology is the study of how individual cells fit within the context of a tissue, what their environment says about their behavior, where they are and why they are there.
Here we talk about the significance of spatial biology in terms of cancer research, with its power to expose how cellular interactions determine the tissue structure and spatial organization within the human body and those implications on disease pathogenesis.
In a recent study from the University of Southern California, researchers used spatial biology to provide novel insights into multiple myeloma. The authors chose a spatial analysis approach to characterize the proteomic profile of myeloma to gain a better understanding about the mechanisms that lead to disease progression and to help identify new biomarkers that can aid clinical diagnostics and therapeutics development.
A little about myeloma
Multiple myeloma is a cancer that starts in the bone marrow as a result of the clonal expansion of resident plasma cells, leading to high tumor burden and organ damage. With an increasing global incidence rate of the disease, there is a strong need to understand the transformation to malignancy and extract relevant molecular signatures.
Changes in plasma cells within the bone marrow are associated with alterations to both their genetic and proteomic profiles, marking a shift from normal to abnormal phenotypes that must be detected with multiple markers in varying combinations.
Monitoring changes in disease using proteomic techniques such as flow cytometry has demonstrated higher applicability when compared with genomic methods for multiple myeloma. However, conventional flow cytometry is low parameter, typically utilizing 4–10 marker panels, and is difficult to standardize with fluctuating panel design and manual analysis. In the midst of these challenges, many alterations to the proteomic landscape of plasma cells during the development, progression, treatment response and disease relapse for multiple myeloma are unknown. Additionally, because the sample type is typically bone marrow, the field is constantly challenged to find ways to analyze these samples with a clarity that offers real insight.
Why high-plex imaging is so helpful
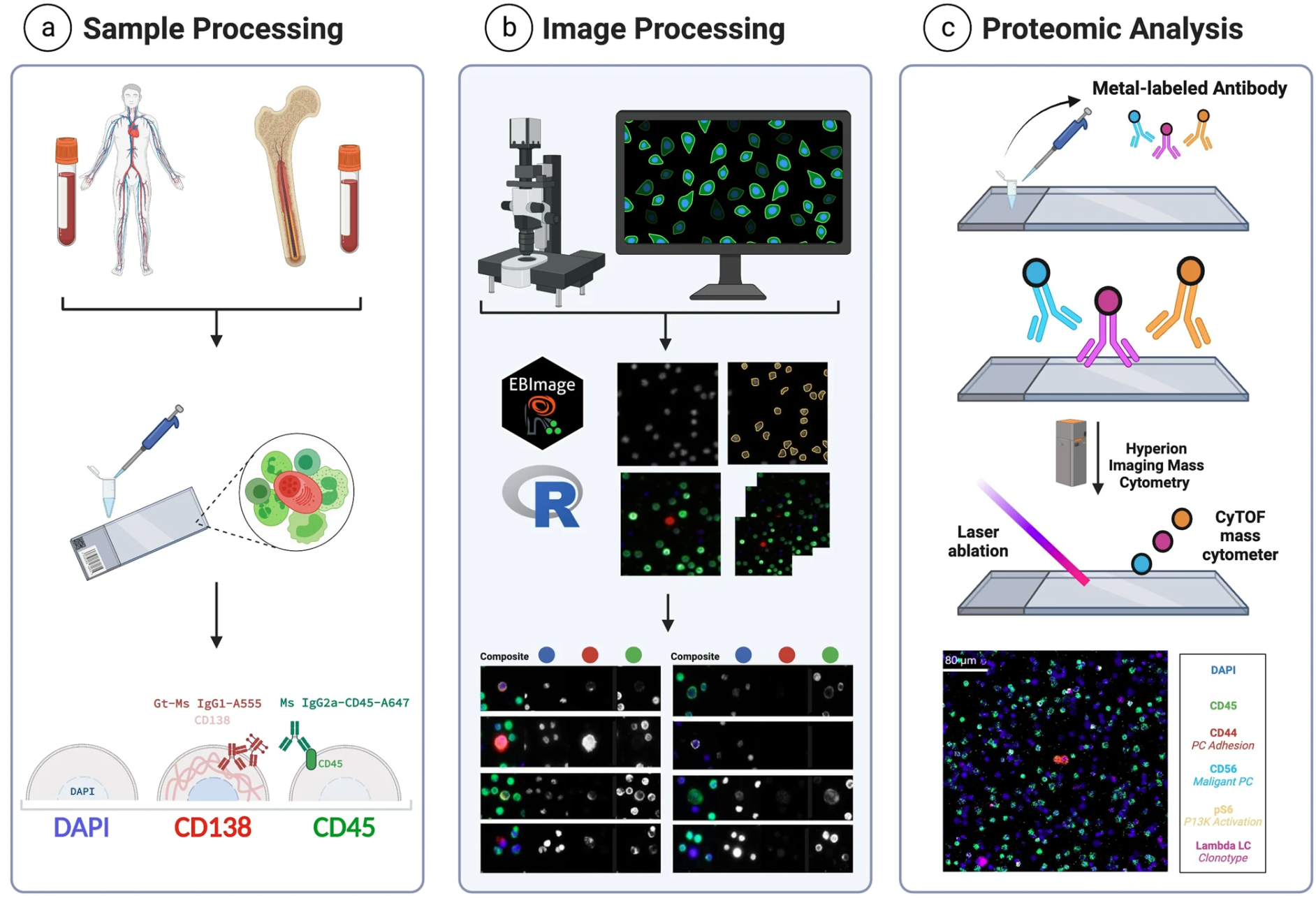
Despite being relatively new, there are several different spatial biology techniques used today that strive to provide the most detailed, highest-resolution, clearest image of what’s going on in a sample. But with so many new technologies out there, it can be tough to figure out which approach best fits with the study goals. For this study, the team chose Imaging Mass Cytometry™ (IMC™) technology.
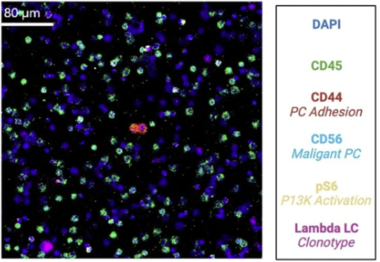
IMC platforms combine cytometry by time-of-flight (on which CyTOF– systems are based) with high-resolution laser ablation to generate spatial information. Tissue sections are stained with panels of antibodies tagged with metal isotopes and scanned, generating isotope plumes that are directed to a mass cytometer for simultaneous analysis. Sequential scanning of 1 µm spots yields a high-dimensional map of target proteins in a tissue or region of interest.
What makes IMC technology unique is the use of metal instead of fluorescent tags, particularly beneficial for samples such as bone marrow. This decreases crosstalk between channels, enabling high-content analysis in a single scan at subcellular resolution. Since IMC workflows leverage time-of-flight mass spectrometry, the technology has the capability to simultaneously stain, acquire and analyze the markers of interest on a tissue section without interference from autofluorescent tissues or management of spectral overlap either in panel design or image analysis.
This challenge of autofluorescence can be a huge limiting factor when staining certain tissue types, such as the brain, lungs, skin, bone marrow and others. This work is an example of how IMC systems can be leveraged to characterize bone marrow cells and profile the heterogeneity of the multiple myeloma tumor microenvironment.
Analysis of bone marrow cells in patient samples across the myeloma disease spectrum using a multiplexed IMC panel was used to characterize the expression of clinical markers for plasma cell classification, potential therapeutic targets and the landscape composition of tumor microenvironment cells. The authors identified four markers [BCMA, ICAM3, CD221 and CS1 (SLAMF7)] as the most abundantly expressed on plasma cells across all myeloma stages, with three of the four (BCMA, ICAM3 and CD221) showing significantly higher expression levels in progressed disease.
The study demonstrates the applicability of spatial biology using multiplexed IMC technology to characterize the proteomic profile of disease for clinical classification, evaluate biomarkers for targeted therapy and provide an overview of the immune landscape all in one assay. This data and the insights gleaned from such comprehensive analysis demonstrate the importance of high-dimensional proteomics to support disease management and reveal signatures with the potential to act as an early predictor of disease development and progression.
Unless explicitly and expressly stated otherwise, all products are provided for Research Use Only, not for use in diagnostic procedures. Find more information here.
