Find your fit: Choosing the right Omics Services platform
Standard BioTools™ Omics Services gives you access to powerful platforms so you can explore molecular, cellular and spatial biology – without the need to purchase instruments or train internal staff.
Whether you’re working in early-stage discovery or clinical research, our technologies support key advances across oncology, neurology, immunology, infectious disease and more. In the sections that follow, we’ll outline what each platform measures and the research contexts where it delivers the greatest value so you can choose the best fit for your study.

SomaScan Platform
High-throughput protein profiling with broad coverage and reproducibility
The SomaScan™ Assay quantifies 11,000 unique human proteins from just 55 µL of serum, plasma or other samples (Table 1). Its scalability, sensitivity and reproducibility make it well suited for longitudinal studies, biomarker discovery and multi-cohort translational research.
Real-world applications
- Illuminates disease biology and informs drug repurposing. In two independent Phase 3 trials of semaglutide to treat obesity and type 2 diabetes, the SomaScan Assay revealed a highly concordant proteomic response across cohorts without the need for bridging samples (Figure 1)1. These insights help clarify disease-related protein expression patterns and may uncover new therapeutic opportunities.
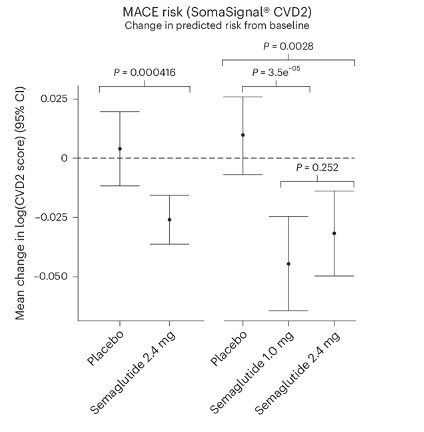
Figure from Maretty et al. (2025).
Figure 1. Effect of semaglutide on predicted cardiovascular disease risk in STEP 1 and STEP 2 participants. The data is presented as mean values with error bars indicating 95% confidence intervals. A two-sided test was used.
- Enables non-invasive diagnostic development. Using the SomaScan Assay, researchers identified a 12-protein plasma signature for hypertrophic cardiomyopathy, validated across eight independent cohorts – all without the need for bridging samples2.
- Tracks disease severity. SomaSignal™ Tests, based on the same technology as the SomaScan Assay, generated consistent protein signatures of non-alcoholic steatohepatitis (NASH) that accurately reflected liver biopsy results and NASH severity, supporting confident longitudinal monitoring3.
- Proven utility in clinical trials. SomaScan technology has been used to:
- Support research models that outperform traditional clinical models in terms of precision, reproducibility, breadth of coverage and detection of biologically relevant protein markers4
- Detect early cardiotoxicity signals, including off-target effects, of the drug torcetrapib5
- Identify immune-related cardiotoxicity in checkpoint inhibitor therapies6
KREX technology
High-specificity antibody profiling for immune-driven biomarker discovery
The KREX™ platform enables high-multiplex antibody profiling, providing deep insight into the adaptive immune response across health, disease and therapeutic intervention (Table 1). It can reveal immunological signals driven by altered proteins, which may play roles in either protective or pathogenic processes. With KREX technology, only correctly folded proteins are available for antibody binding, ensuring high specificity and reproducibility.
Real-world applications
- Identifies predictive immune signatures. The KREX platform identified a 13-autoantibody survival signature following surgical resection of early-stage non-small-cell lung cancer (NSCLC)7. The model showed 84–89% sensitivity and 74–80% specificity across both discovery and validation cohorts.
- Differentiates disease subtypes for targeted drug development. In systemic lupus erythematosus (SLE), the KREX platform revealed 79 SLE-discriminating autoantibodies and four clusters of functionally linked autoantigens that could potentially help predict organ involvement and guide patient stratification8.
CyTOF technology
Single-cell immune profiling with deep marker resolution and clinical scalability
Mass cytometry, powered by CyTOF™ technology, enables high-parameter immune cell profiling at the single-cell level – capturing over 50 markers simultaneously with no spectral overlap. This allows for a comprehensive view of immune cell function and phenotype (Table 1). CyTOF technology has been widely adopted in translational and clinical research, including over 100 clinical trials, and is especially well suited for studies in which immune modulation plays a key role.
- Supports patient stratification based on immune profiles. In a Phase 2 study in endometrial cancer (EC), a 36-marker CyTOF panel was used to analyze baseline tumor biopsies. The resulting data identified immune cell subsets that stratified patients as progressors or non-progressors to nivolumab and cabozantinib, becoming the first reported biomarkers of immunotherapy response in EC.
- Reveals immune signaling dynamics in aging. In another Phase 2 trial, CyTOF technology was used to analyze immune responses in older patients undergoing primary hip or knee arthroplasty. Treatment with young plasma proteins led to measurable shifts in both proteomic and intracellular signaling profiles, pointing to mechanisms by which young plasma modulates age-associated immune pathways to surgical trauma.
- Enables multi-center clinical trials. Stable CyTOF reagents simplify logistics across study sites and preserve data quality, ensuring high reproducibility even when sample collection and processing occur at different locations or times.
Hyperion systems
High-plex spatial proteomics for quantitative tissue-based insights
Hyperion™ Imaging Systems, built on Imaging Mass Cytometry™ (IMC™) technology, enable simultaneous detection of 40-plus markers within intact tissue architecture for spatial biology insights (Table 1). Unlike traditional immunohistochemistry (IHC), this approach delivers quantitative, high-resolution imaging on a continuous scale, providing an objective foundation for patient stratification and increasing statistical power in tissue-based studies.
Why it matters: A breast cancer case study
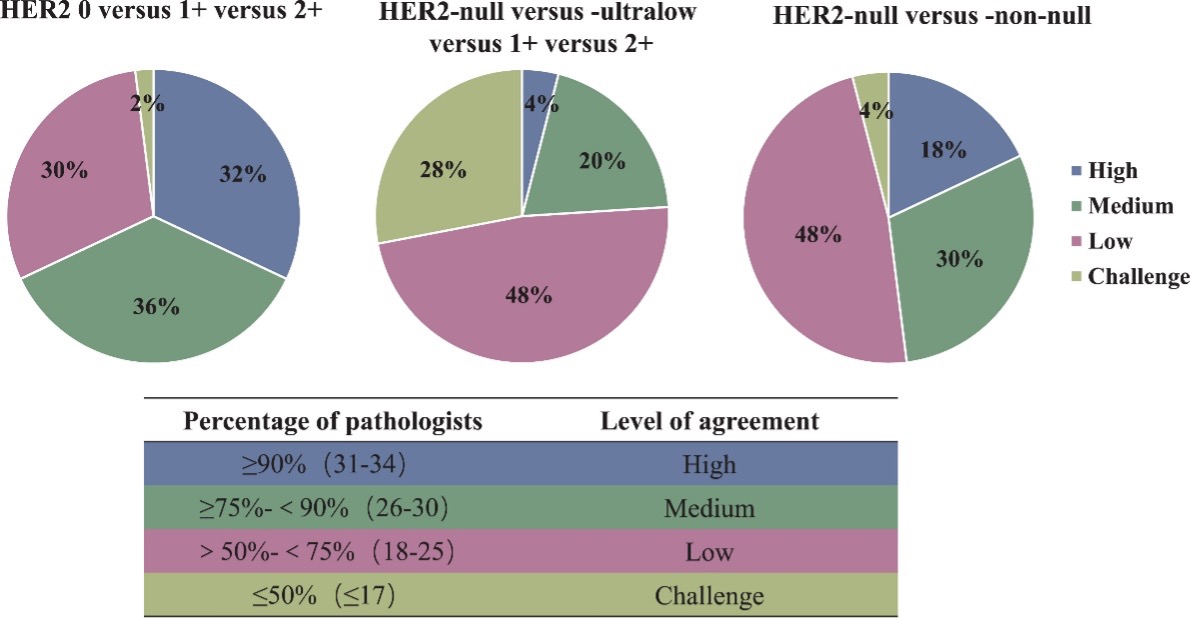
Figure from Wu et al. (2025).
Figure 2. Assessment of interpretation consistency for each case across different HER2 categories. Based on HER2 scores provided by 36 pathologists, cases were classified into three categories: HER2 IHC 0, IHC 1+ and IHC 2+. Of these cases, 32% (16/50) demonstrated high consistency, 36% (18/50) showed moderate consistency, 30% (15/50) had low consistency and 2% (1/50) were classified as challenging.
- IHC cannot reliably detect low HER2 expression. Pathologists assess HER2 – a key biomarker in breast cancer – with IHC scoring and reflex in situ hybridization to determine treatment. However, patients with HER2-low and -ultralow expression were marked as faint, incomplete or barely perceptible, leading to inconsistent and inaccurate patient stratification, and posing diagnostic challenges9.
- IHC resulted in poor inter-observer consistency. There is low diagnostic concordance among pathologists using IHC when evaluating HER2-ultralow breast cancer samples (Figure 2). This inconsistent differentiation results in unreliable patient stratification10.
- Hyperion systems offer a more precise alternative. IMC platforms provide a continuous scale of expression, enhanced contrast resolution and greater dynamic range than IHC. They enable objective quantification of biomarkers, overcoming the variability of categorical scoring. IMC technology can also be applied to existing or new IHC slides, allowing for comprehensive tissue reanalysis or expansion of retrospective cohorts.
Conclusion
Selecting the right platform is essential to unlocking meaningful insights in translational research. Our Omics Services offers end-to-end support – from study design through data analysis – leveraging world-class platforms to explore biology at molecular, cellular and spatial levels. With SomaScan, KREX, CyTOF and Hyperion technologies, researchers can advance precision medicine by revealing protein signatures that provide a fundamental understanding of disease to aid in the development of better disease detection, risk assessment, monitoring capabilities and therapeutic response. These biomarkers help de-risk development and translate data into impact.
Table 1. Standard BioTools Omics Services platforms
Platform |
Measured Analyte |
Multiplexing Capability1 |
Sample Type |
Application Strengths |
|---|---|---|---|---|
| SomaScan Assay, SomaSignal Tests | Protein | 10,000-plus | Serum, plasma and other biological fluids2 | Broad proteomic screening for biomarker discovery; cross-cohort comparability; longitudinal monitoring in translational and clinical studies |
| KREX | Antibody | 1,800-plus | Serum, plasma and other biological fluids2 | High-specificity antibody profiling; immune signature discovery in health, disease and therapeutic response; molecular subtyping and risk stratification |
| CyTOF | Protein | 50-plus | Cells, including whole blood | Deep immune cell phenotyping; patient stratification; single-cell analysis for treatment response and mechanistic studies |
| Hyperion Imaging System | Protein | 40-plus | Tissue | Spatial biomarker mapping; links drug exposure to clinical outcomes in tissue-based research; reanalysis of archival slides for retrospective studies |
- The number of analytes analyzed simultaneously in a single assay. Lower-plex panels are also available.
- Other biological fluids may require optimization.
Ready to move forward with your research project?
References
- Maretty, L. et al. “Proteomic changes upon treatment with semaglutide in individuals with obesity.” Nature Medicine 31 (2025): 267–277.
- Wang, Z. et al. “Development of a blood-based multiple-marker screening test for hypertrophic cardiomyopathy.” Journal of Cardiac Failure 30 (2024): S7.
- Sanyal, A.J. et al. “Defining the serum proteomic signature of hepatic steatosis, inflammation, ballooning and fibrosis in non-alcoholic fatty liver disease.” Journal of Hepatology 78 (2023): 693–703.
- Williams, S.A. et al. “A proteomic surrogate for cardiovascular outcomes that is sensitive to multiple mechanisms of change in risk.” Science Translational Medicine 14 (2022): eabj9625.
- Williams, S.A. et al. “Improving assessment of drug safety through proteomics: Early detection and mechanistic characterization of the unforeseen harmful effects of torcetrapib.” Circulation 137 (2018): 999–1010.
- Mohindra, R. et al. “1173 Proteomic analysis in patients with immune checkpoint inhibitor (ICI) associated cardiotoxicity.” Journal for ImmunoTherapy of Cancer 12 (2024).
- Patel, A.J. et al. “A highly predictive autoantibody-based biomarker panel for prognosis in early-stage NSCLC with potential therapeutic implications.” British Journal of Cancer 126, 2 (2022): 238–246.
- Lewis, M.J. et al. “Autoantibodies targeting TLR and SMAD pathways define new subgroups in systemic lupus erythematosus.” Journal of Autoimmunity 91 (2018): 1–12.
- Sajjadi, E. et al. “Improving HER2 testing reproducibility in HER2-low breast cancer.” Cancer Drug Resistance 5 (2022): 882–888.
- Wu, S. et al. “Interobserver consistency and diagnostic challenges in HER2-ultralow breast cancer: a multicenter study.” ESMO Open 10 (2025): 104127.
The SomaScan, KREX, CyTOF and Hyperion platforms are for Research Use Only. Not for use in diagnostic procedures.
