The significance of the messenger (RNA)
How we can tackle the complexity of cell signaling and translate this knowledge to what matters
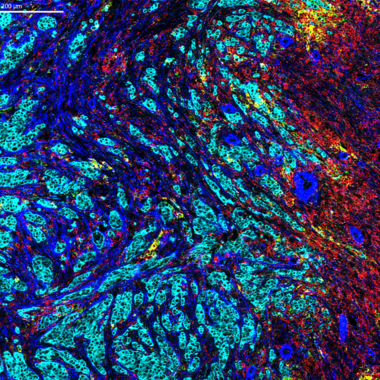
Our days are full of complex tasks – not completed by us but accomplished through the interactions of the body’s trillions of cells to enable our higher function. These critical activities help us maintain ourselves, or any multicellular organism, and are done through cell signaling.
Cell signaling – how a cell exchanges messages with its environment and itself – underlies several processes such as cell growth, differentiation, migration and apoptosis. When cells communicate with each other, complex signaling pathways comprised of small molecules and proteins such as chemokines, hormones or neurotransmitters must work to their fullest capacity.
Perturbation of these signaling pathways has been associated with the development of a variety of diseases due to mutations or abnormal expression of signaling components. Cell signaling aberrations in cancer can occur when cells ignore their regular signals to stop proliferating. In immune disorders, immune cells can get inappropriately activated through rampant cell signaling and stimulate an exaggerated immune reaction at the wrong target, such as our own cells. During an infection, even pathogens can exploit cell signaling pathways to benefit their own survival.
Deciphering how cells communicate with each other and themselves gives us a clearer look into the mechanisms of disease and can ultimately help develop new treatments that target signaling molecules. Gaining a better understanding of cell signaling can also help in assessment of drug response. However, the complexity of signaling pathways makes it difficult to know if a treatment will improve health outcomes while avoiding detrimental and hard-to-predict side effects. In this broader context, knowledge of individual pathways must be extended to consider a wider range of interactions as a whole. Better prediction of outcome or off-target effects, for example, requires this approach of examining all levels of signaling capacities and connections.
Being able to use a multi-omic approach to study cell signaling pathways helps determine the planned versus actual activity occurring within the cells. For imaging techniques, visualizing RNA and protein markers in the same sample helps provide more of that crucial understanding. Proteins, the functional unit translated from messenger RNA, are variable in size, shape and function and can be subject to post-translational modifications that make them more difficult to detect. It can be extremely challenging to generate antibodies for such proteins, or other small secreted proteins, integrated membrane proteins or rapidly degraded proteins. Adding RNA detection enables researchers to visualize these harder-to-capture proteins and gain more in-depth information about what’s really happening within cells.
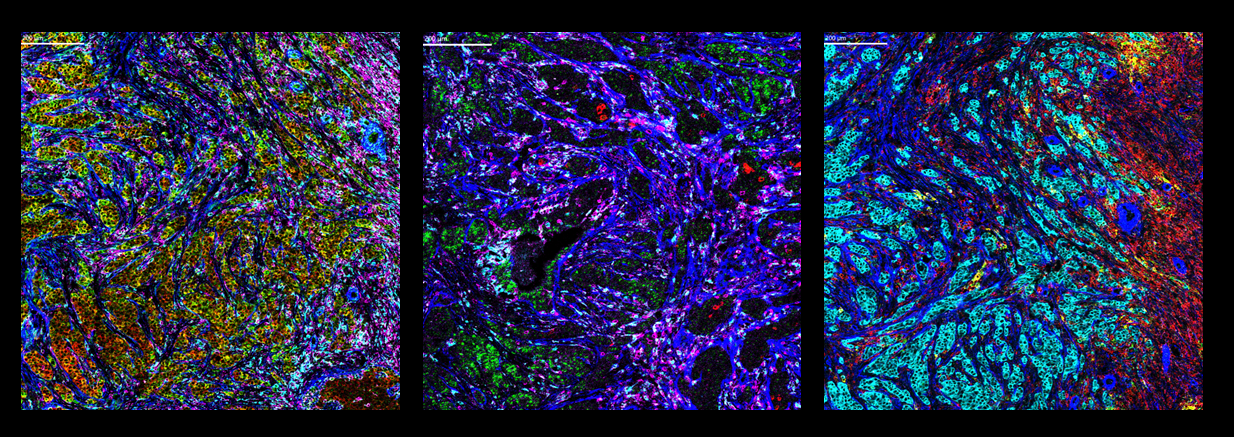
Left: Co-detection of positive control RNA and proteins on FFPE human breast cancer tissue. Positive control RNA GAPDH and ACTB along with protein targets αSMA, CD45 and CD68 are displayed.
Center: Co-detection of target RNA and proteins on FFPE human breast cancer tissue. RNA targets CXCL13 and VEGFA along with protein targets αSMA, CD45 and CD68 are displayed.
Right: Co-detection of target RNA and proteins on FFPE human breast cancer tissue. RNA targets CXCL9 and CXCL10 along with protein targets αSMA, pan-keratin and CD45 are displayed.
A new workflow employing Imaging Mass Cytometry™ and RNAscope™ technology allows researchers to detect all key targets in a disease-perturbed signaling pathway in this manner. Using the workflow, samples from a control group and a drug-treated group (for example, individuals who received a placebo versus individuals who received a signal-pathway-altering drug) can be compared, and researchers can apply data collected about a particular signaling event to determine if drug treatment altered that pathway. This ability to simultaneously detect RNA and protein allows researchers to gain these deeper insights into complex cellular interactions through the inclusion of targets that they might not have otherwise been able to study.
Learn more about how to leverage the subcellular resolution and multiplexing capabilities of IMC™ with the high sensitivity and specificity of RNAscope to study previously inaccessible targets and activation states of cells and drive further biological insights. If you want to learn more about utilizing RNAscope and protein co-detection, check out this GEN webinar from Sammy Ferri-Borgogno on using a multi-omic approach to tissue imaging to reliably characterize the tumor microenvironment in ovarian cancer.
Check out other blog posts
Harness the power of mouse tumor models
How to enable studies that reveal significant details of tumor development, progression and treatment.
Bringing the CyTOF advantage to CAR cell therapy research
Standard BioTools shares how new advancements in immunophenotyping are accelerating CAR cell therapy development and improving precision medicine.
Unless explicitly and expressly stated otherwise, all products are provided for Research Use Only, not for use in diagnostic procedures. Find more information here.



